RECENTLY READ
UPCOMING READING
3D U-Net Learning Dense Volumetric Segmentation from Sparse Annotation
Jan 3, 2021 DL 3D U-Net Segmentation
Table of Contents
Summary
Çiçek et al. extended the idea of the u-net to 3D images. The kernels of the network were adjusted to the 3D by adding an additional dimension to the kernel size.
The network learnes in to possibilities, once from user annotated slices or fully automated from a annotated training set.
The images were of Xenopus kidneys.
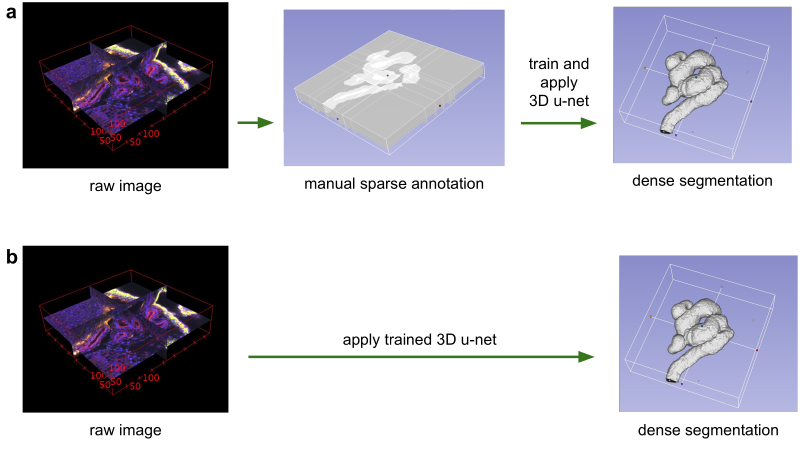 Application scenarios for volumetric segmentation with the 3D u-net.
(a) Semi-automated segmentation: the user annotates some slices of each vol-ume to be segmented.
The network predicts the dense segmentation. (b) Fully-automated segmentation:
the network is trained with annotated slices from arepresentative training set
and can be run on non-annotated volumes.
Application scenarios for volumetric segmentation with the 3D u-net.
(a) Semi-automated segmentation: the user annotates some slices of each vol-ume to be segmented.
The network predicts the dense segmentation. (b) Fully-automated segmentation:
the network is trained with annotated slices from arepresentative training set
and can be run on non-annotated volumes.
Network
Like the original u-net the network consists of two paths.
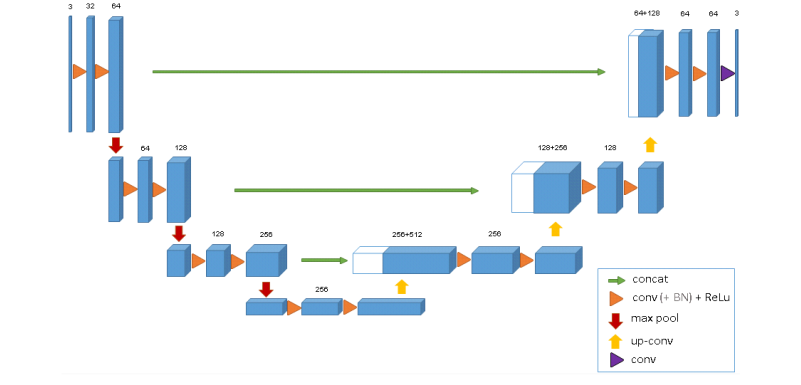 The 3D u-net architecture. Blue boxes represent feature maps.
The num-ber of channels is denoted above each feature map.
The 3D u-net architecture. Blue boxes represent feature maps.
The num-ber of channels is denoted above each feature map.
In this network also used the batch normalization after every ReLU layer. The authors claim the loss function played an important part in the network:
The important part of the architecture, which allows us to train on sparse annotations, is the weighted softmax loss function. Setting the weights of unla- beled pixels to zero makes it possible to learn from only the labelled ones and, hence, to generalize to the whole volume.
Data Augmentation
To augment the data rotation, scaling and gray value augmentation was used. Additionally a smooth dense deformation field on both data and ground truth labels was applied. For this, the authors choose random vectors applied a B-spline interpolation.
Metadata:
| Author: | Özgün Çiçek, Ahmed Abdulkadir, Soeren S. Lienkamp, Thomas Brox, Olaf Ronneberger |
| Released: | Jun 21, 2016 |
| Source: | Link |